Spatial Collaborative Services
Additional services are in development. For questions, please email [email protected].
10X Genomics Visium v1
The Visium spatial transcriptomics platform enables whole tissue, spatially resolved, gene expression profiling in both fresh frozen and FFPE material. The system provides transcriptomic information within the context of tissue architecture, tumor microenvironments and cell groups.
The platform relies on custom Visium slides, with either two or four capture areas upon which tissue sections are affixed. Capture areas are either 6.5 x 6.5 mm with 5000 tiled capture spots or 11 x 11 mm with 14,00 capture spots. Each spot has an average capture radius of 50 uM and contains millions of oligonucleotides with a polyA capture sequence, a unique molecule identifier (UMI) and an indexed spatial barcode. After capture, Illumina-sequencing compatible libraries are generated from the probe products and used for high-throughput sequencing.
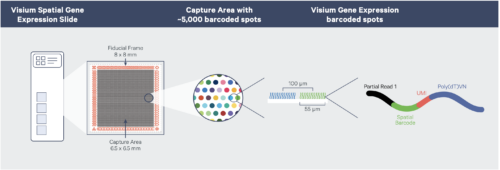
10X Visium v1 workflow (courtesy of 10x Genomics)
10X Genomics Visium HD
The VisiumHD spatial transcriptomics platform expands on the Visium v1 platform to enable whole tissue, spatially resolved gene expression in FFPE material. The system provides higher-resolution transcriptomic information with no dropout areas, allowing for near-single-cell assessment of tissue architecture, microenvironments and co-expression networks.
The assay relies on Visium HD slides containing two capture areas, each 6.5 x 6.5 mm. Capture areas are arrayed with a continuous 2×2 uM lawn of barcoded oligonucleotides. The assay utilizes probe-based gene expression chemistry; probes are hybridized to H&E- or IF-stained FFPE material on a glass slide and imaged to capture spatial orientation. The 10x CytAssist instrument then is used to transfer the transcriptomic probes to the Visium HD slide and extended to add location-specific barcodes. After capture, Illumina sequencing compatible libraries are generated from the probe products and used for high-throughput sequencing.
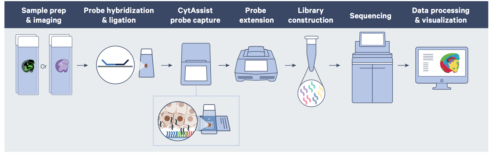
Source: 10x Genomics Grant application Resources for Visium Spatial Gene Expression
10X Genomics Xenium
Xenium is a high-resolution, imaging-based spatial profiling system that can characterize up to 5,000 genes and multiplexed proteins in the same tissue section. This system combines spatial fidelity with subcellular resolution in FFPE and fresh frozen tissue, facilitating experiments desiring detailed cell type characterization or exploring cell-cell interactions and transcriptional pathways within their biological contexts. Tissues can be assayed using pre-designed, validated disease or tissue-focused gene panels, fully customized panels, or a mix of the two.
Processing starts with tissue immobilized on a custom Xenium slide, which contains 10.5 x 22.5 mm of surface area to assay multiple tissue sections at a single time; up to two slides can be prepared per run. Probes are hybridized to RNA in the tissue, amplified, and sequenced via cyclic fluorescent images taken at a 200nm per-pixel resolution. Data is visualizable via desktop software from 10X or third parties to localize RNA and protein signals and perform secondary analysis.
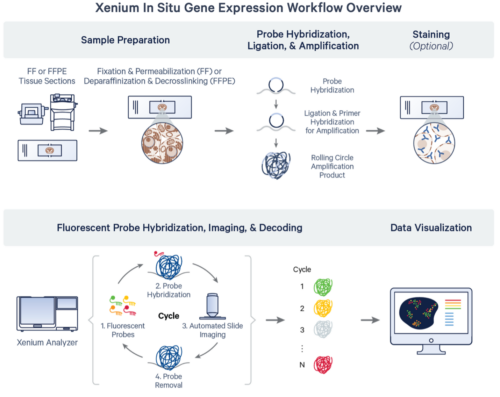
Source: 10x Genomics Xenium In Situ Gene Expression Protocol Planner (CG000601 rev F)
Nanostring GeoMx DSP
The GeoMx DSP enables spatially resolved, region of interest targeted, profiling of both mRNA and proteins in both fresh frozen and FFPE tissue.
The assay is performed on standard negatively charged slides upon with the tissues of interest are mounted; antibodies or RNA probes coupled to photo-cleaveable barcode oligos are incubated with both the tissue and morphology markers. After staining, regions of interest (ROI) can be chosen and optionally subsectioned based on morphology. Oligos from the selected regions are then released by exposure to UV light, incorporated into an Illumina sequencer compatible library and used for high throughput sequencing.
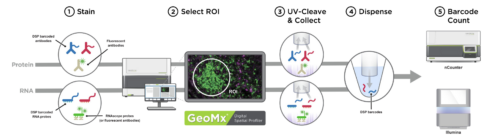
Nanostring GeoMx DSP workflow (Courtesy of Nanostring)
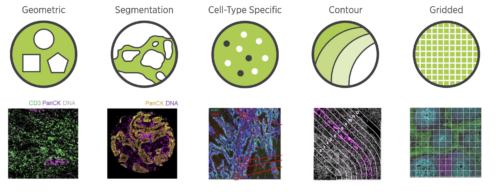
Available Nanostring Region of Interest (ROI) segmentation options (courtesy of Nanostring)
What are the differences between spatial assays?
| Platform | Sample Type | Submission Format | Presubmission QC | Imageable area | Species Compatibility | RNA Compatible | Protein Compatible | Regions Assayed | Resolution | Detection | Deliverables |
| 10x Genomics Visium v1 | Fresh Frozen or FFPE | Blocks or prepared 10x custom slides | RIN/DV200 score: Fresh = RIN >7.0; FFPE = DV200>50% | 6.5 mm x 6.5 mm. 4 regions/ one time use slide | Fresh Frozen = All eukaryotes; FFPE = Human/Mouse | Yes | FFPE only | Whole slide | ~55uM diameter (1-10 cells) | NGS | Fastq files appropriate for Space Ranger or third party pipeline |
| 10X Genomics Visium HD | Fresh Frozen, Fixed Frozen, or FFPE | Blocks or freshly cut charged slides | FFPE = DV200>50% | 6.5 mm x 6.5 mm 2 regions/slide | Human/Mouse | Yes | No | Whole slide | 2×2 uM grid | NGS | Fastq files appropriate for Space Ranger or third party pipeline |
| 10X Genomics Xenium | FFPE | Blocks or prepared 10X custom slides | FFPE = DV200>50% | 10.5 x 22.5 mm/slide | – Predesigned panels: Human/mouse – Custom Panels: species agnostic | Yes | Yes | Whole slide | Subcellular: Transcript XY-localization precision < 30 nm and Z-localization precision < 100 nm Pixel size = ~0.2 µm/pixel | Imaging | Cell Feature matricies, Transcripts (CSV), Tissue Images |
| Nanostring GeoMx DSP | Fresh Frozen, FFPE, Fixed Frozen | Blocks or freshly cut charged slides | None | 35 mm x 14 mm | Human/Mouse | Yes | Yes | Targed regions of interest, user defined | 200-600uM typical (50-100 cells) | Fastq + DSP localization files |
Frequently Asked Questions
How do I start a project?
Please fill out this form or click the button below and a core member will be in touch to schedule an initial planning meeting with the Spatial group, which includes individuals from the Genomics, Histology, Imaging, and Informatics.
What services can be included in a spatial transcriptomics experiment?
- Embedding/re-embedding of fresh frozen/FFPE samples
- Sample quality checks (RIN/DV200 scores, H&E pathology review)
- Sectioning and section placement
- Optimization for permeabilization or antibody times
- Morphology checks (GeoMx only)
- Expression analysis
This includes imaging, slide work, library preparation, sequencing, and basic data analysis.
Questions about 10X Visium
What types of samples can be used in 10X Visium Experiment?
The following samples can be used in a 10X Visium experiment:
- Fresh frozen samples of human or animal origin
- FFPE samples of either mouse or human origin
How should samples be collected?
Fresh frozen samples: Flash freeze in liquid nitrogen cooled isopentane as soon as possible after collection. Optimally embed in OCT or CMC either at the same time or afterward. Tissue sections should be <6.5mm x 6.5mm to be compatible with Visium spatial slides. Optimal tissue thickness is 10um. High fat content tissues may require thicker sections.
Fresh Frozen Tissue Prep Protocol
FFPE samples: Please follow standard formalin fixation and embedding protocols. Samples should ideally be stored at 4°C post fixation to preserve RNA quality. Tissue sections should be <6.5mm x 6.5mm or <11mm x 11mm to be compatible with Visium spatial slides. Optimal tissue thickness is 5 um.
What are recommended sample quality control steps?
Core support is available for all QC steps listed below.
Fresh frozen:
- RNA extraction (RIN >7.0) on 10 uM section
- Visium Optimization Experiment to assess permeabilization times
Repository stored FFPE:
- RNA extraction (DV200 >50%) and H&E stain on 5 uM sections. NOTE: FFPE done on fresh samples by VAI’s Histology team do not need this step
- Visium Tissue Adhesion Test to assess optimal hydration times
What is the resolution of 10X Visium?
- 55 um capture spots, 100um center to center. Each spot will capture 1–10 cells, depending on tissue type; each spot should not be thought of as a single cell.
- 5000 spots per 6.5mm x 6.5mm capture area.
- 14,000 spots per 11mm x 11 mm capture area.
- Entire section is captured at the same time.
Types of Visium slides
Fresh frozen: 6.5mm x 6.5mm, 4 samples per slide

FFPE with CytAssist: 6.5mm x 6.5mm or 11mm x 11mm, 2 samples per slide
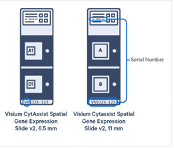
Question about Nanostring GeoMx DSP
What types of samples can be used in a Nanostring GeoMx DSP Experiment?
The following samples can be used in a Nanostring GeoMx DSP experiment: FF or FFPE, OCT from mouse or human tissues only.
How should samples be collected?
- Fresh frozen samples: Flash freeze in liquid nitrogen cooled isopentane as soon as possible after collection. Optimally embed in OCT or CMC either at the same time or afterward.
- FFPE samples: Please follow standard formalin fixation and embedding protocols. Samples should ideally be stored at 4°C post fixation to preserve RNA quality.
Optimal tissue thickness is 5um.
Tissue should be sectioned onto positively charged slides no more than two weeks prior to running the Nanostring DSP assay.
What are recommended sample quality control steps?
Morphology check: A test slide for a scanning only run (without ROI definition, UV illumination, or ROI collection) is strongly recommended to expedite ROI selection the day of the experiment and validate the fluorescent staining of your morphology markers. All requests utilizing custom antibodies are required to undergo this process.
What is the resolution of GeoMx DSP?
Minimum of 50–100 cells per ROI (region of interest). May be subdivided into multiple AOIs (areas of interest) based upon morphology markers in that region.
What are the recommended slides and configuration?
Multiple sections or sections from multiple tissues can be placed on the same slide, so long as the same probe set will be used on all slides.
All sections must fit within the 40mm x 17mm slide region and be entirely within the GeoMx gasket to ensure a good seal.
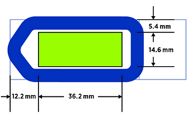
Which slides are compatible?
- Leica Bond Plus (Cat# S21.2113.A)
- Fisherbrand Superfrost Plus (Cat # 12-550-15)
Which panels are available?
- Whole transcriptome atlas (human or mouse)
- Cancer transcriptome atlas (human)
- Protein assays with NGS readout (human or mouse)
