Research brief: Developing a “roadmap” for the second deadliest cancer worldwide
June 20, 2017
Epigenetics: Refers to the packaging and marking of DNA, which guides whether genes are on or off and to what extent. Epigenetic errors can lead to diseases such as cancer.
Exome: The total number of expressed genes in a cell.
Gene: The smallest unit of heredity, genes are sequences of DNA that hold instructions for specific functions. Genes can be thought of as recipes for life, determining everything from eye color to disease risk.
Gene expression: The process by which instructions encoded in a gene are acted upon.
Gene mutation: A permanent change to a gene that may affect its function. For example, a mutation may prevent a cell from knowing when to stop replicating, which may lead to cancer. Read more here.
Hepatitis: A disease marked by inflammation of the liver, most commonly caused by a virus. Read more here.
Oncogene: A gene that can cause cancer, under certain conditions.
Tumor suppressor: A gene that acts as a guardian for the genetic code, by ensuring that cell division occurs appropriately and that mistakes in the DNA are fixed, among other tasks. Cancer can arise when these genes are inappropriately silenced, or made inactive.
Scientists have a new tool to tackle the second leading cause of cancer death, thanks to work from The Cancer Genome Atlas Research Network, a National Institutes of Health-funded, multi-institutional team dedicated to molecularly mapping cancers.
Their findings, published last week in Cell, are the first large-scale, multi-platform analysis of its kind in liver cancer, a major public health threat that claims more than 700,000 lives annually worldwide. Few effective treatments exist and prognosis is generally poor, with only 31 percent of patients with localized disease surviving five years after diagnosis. That number drops to 11 percent for people with regional disease, meaning the cancer has spread to nearby areas, and falls to only 3 percent for people whose cancer has spread to distant sites in the body.
“We hope that these findings, which represent the most comprehensive analysis of liver cancer to date, will lead to new avenues of research and better stratification of people in clinical trials—both important components to developing more effective treatments,” said Dr. Peter W. Laird, a professor at Van Andel Research Institute who led the project’s epigenetic analysis. “Studies such as this really demonstrate the power of collaboration. Without it, a project of this scope would not be possible.”
The study comes at a critical juncture. Since 1980, liver cancer incidence has tripled, due in part to rising rates of diabetes and obesity. Other risk factors include chronic hepatitis B or C infections, alcohol abuse and exposure to some environmental agents, such as aflatoxin B1, a toxin produced by certain molds that grow in hot, humid climates. Each of these can damage the liver, causing an inflammatory cascade that interferes with the normal cellular life cycle and damages the genetic instructions responsible for ensuring healthy function.
“Our findings showcase the incredible complexity of liver cancer,” said Dr. Toshinori Hinoue, a bioinformatics scientist in Laird’s lab who conducted the epigenetic analysis. “While this presents a significant challenge, it also provides an outstanding opportunity to develop new ways to precisely target subsets of the disease, rather than a one-size-fits-all approach.”
Here are some of the team’s findings, distilled down to the major points (read the full paper here for all of the details).
Genetic factors
At its core, cancer is a genetic disease, prompted by mix-ups in the code that contains the instructions for life. The body has systems in place to prevent these errors and to fix them when they occur, but occasionally they slip by and prompt the uncontrolled, invasive cell growth that is cancer’s hallmark.
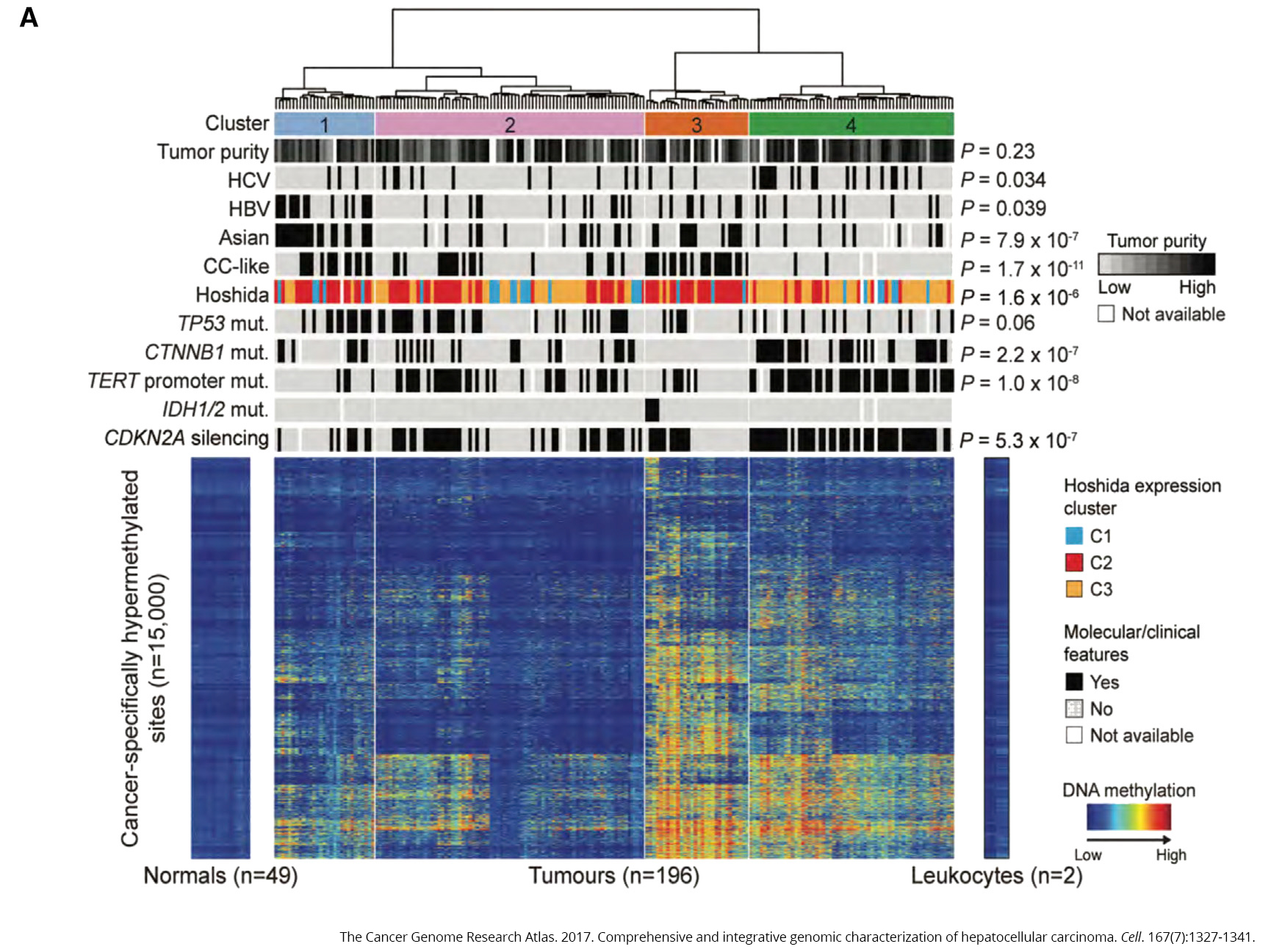
To see what was happening behind the scenes in liver cancer, The Cancer Genome Atlas team looked at the full exomes (the total set of expressed genes in a cell) of 363 cases. They found 12,136 genes with mutations across the genome. Of those, 26 were found to significantly mutated, including genes that suppress tumors (TP53, AXIN1 and RB1), the oncogene CTNNB1 and in genes that help package DNA (ARID1A, ARID2 and BAP1), to name a few.
The most common mutation was in a gene called TERT, which contains the instructions for telomerase, an enzyme that acts as a maintenance worker for the telomeres that comprise the ends of chromosomes. Telomeres can be thought of as biological timers, becoming shorter each time a cell divides. Once they become short enough, they stop the cell from dividing and start the process of cell destruction. Without this process, cells would keep dividing and accumulating errors—a situation that often leads to cancer.
Moreover, TERT mutations were linked to silencing of CDKN2A, a tumor suppressor gene that helps ensure a cell’s life cycle runs smoothly. When CDKN2A gets shut down, cells no longer know when to die, allowing them to continue replicating, regardless of the mistakes and damage they may have accrued.
Lastly, 31 percent of tumor samples had mutations in TP53, which is one of the most commonly mutated genes across all cancers. This gene is the subject of intense research efforts to find ways to fix mutations in it, ultimately getting its ability to regulate the cell cycle back on track.
Epigenetic errors
While genetic mutations refer to changes to the genetic code itself, epigenetic changes, or modifications as they’re more commonly called, refer to that the processes used to read and express the code.
One of the most common epigenetic modifications is called DNA methylation, during which complexes called methyl groups are added to or removed from the DNA. This process helps activate or silence genes.
To see how methylation may play a role in the disease, the team compared samples from healthy liver tissue to 196 samples from people with liver cancer. What they found was striking—the cancer samples exhibited significant variation in methylation, with too much in some places and too little in others (caller hyper- and hypomethylation, respectively).
Two of the genes that experienced too much methylation were IDH1 and IDH2, oncogenes that have been linked to problems with cellular metabolism. It’s not the first time these genes have cropped up in a TCGA study—earlier this year, the team found that mutations in these genes marked one of four subtypes of choloangiocarcinoma, a rare bile duct cancer that shares many of the same features as liver cancer. Differentiating between these two diseases is an important—and sometimes extremely difficult—component of disease diagnosis.
Viral infections
Chronic infection with either hepatitis B or hepatitis C viruses have long been known to increase a person’s risk for developing liver cancer. About 22 percent (44 of 196 samples) had evidence of hepatitis B infection versus 17.9 percent (35 of 196 samples) for hepatitis C.
In the majority of samples from people with hepatitis B, DNA from the virus had integrated with the person’s own DNA, leading to the idea that this could interfere with the function of tumor suppressor genes. In hepatitis C samples, on the other hand, the tumor suppressor gene CDKN2A was more frequently silenced and TERT mutations were more common (remember those from earlier?).
The role of the immune system
In addition to molecular techniques, the team also utilized histopathology to tease out the types of cells present in 196 tumor samples. About 22 percent had medium to high levels of immune cells and molecules related to the immune system. Harnessing this system, which acts as the body’s natural defenses, is a prominent area of cancer research and may help sensitize tumors to additional drug therapy.
The Cancer Genome Atlas is a National Institutes of Health-funded, multi-institutional effort to molecularly map cancers of significant public health importance. VARI authors include Dr. Peter W. Laird, who led the epigenetic analysis, Dr. Toshinori Hinoue, Dr. Hui Shen and Dr. Stephen Baylin (whose primary appointment is at Johns Hopkins University). The liver cancer project was led by Dr. David A. Wheeler of Baylor College of Human Medicine and Dr. Lewis R. Roberts of Mayo Clinic.
